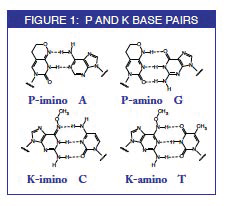
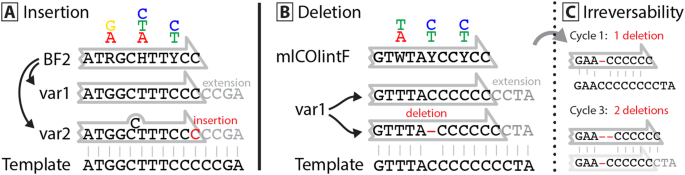
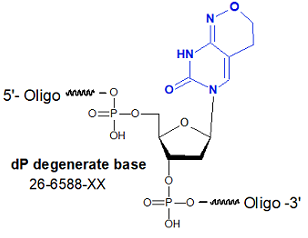
Degenerate primer design. Degenerative Wnt primers (designated Upper... | Download Scientific Diagram
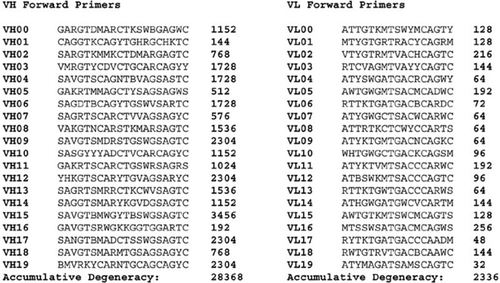
Skip the Alignment: Degenerate, Multiplex Primer and Probe Design Using K-mer Matching Instead of Alignments | PLOS ONE

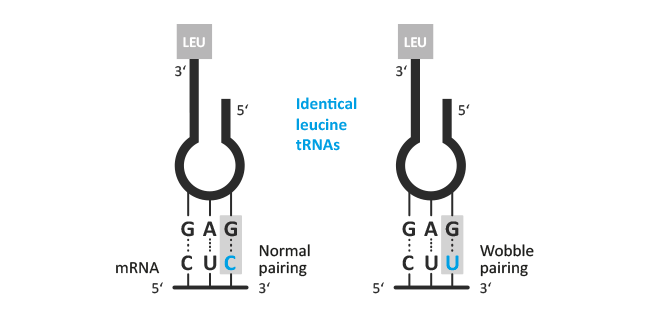
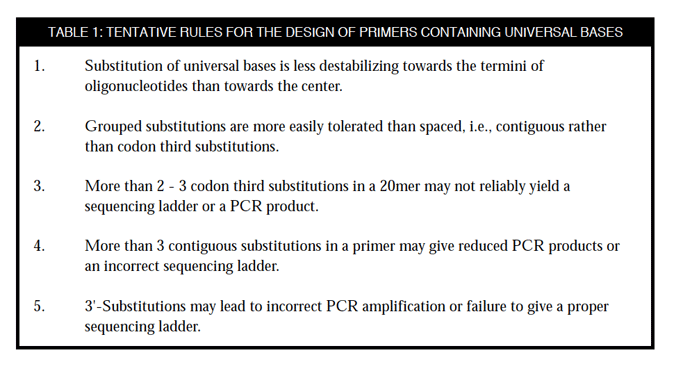
SOLVED: These primers are known as “degenerate” primers because there is variation in the bases present at certain sites. Why might it be advantageous to include degenerate sites in the primers?

Design of degenerate PCR primers. (A) Alignment of six mammalian L1... | Download Scientific Diagram

Development of RT-PCR degenerate primers for the detection of two mandariviruses infecting citrus cultivars in India - ScienceDirect
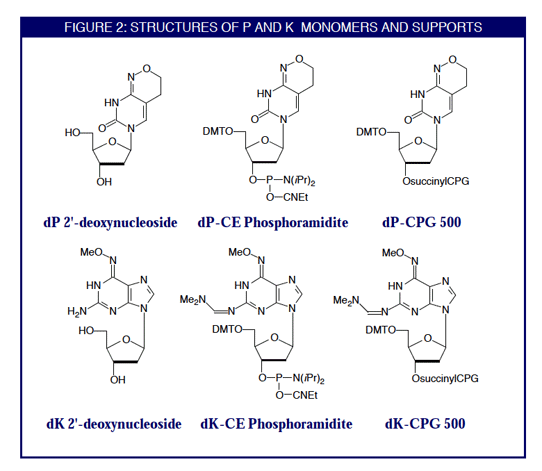
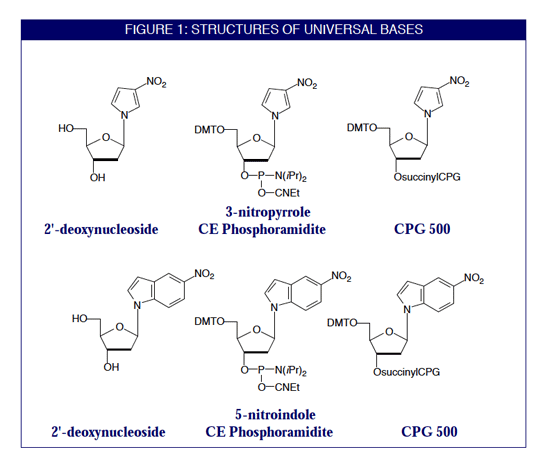
![PDF] Degenerate Primer Design Using Computational Tools Computational Molecular Biology | Semantic Scholar PDF] Degenerate Primer Design Using Computational Tools Computational Molecular Biology | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/89cb6d6c927e29073c9fd66ccfab088d2f9895ff/10-Figure4-1.png)
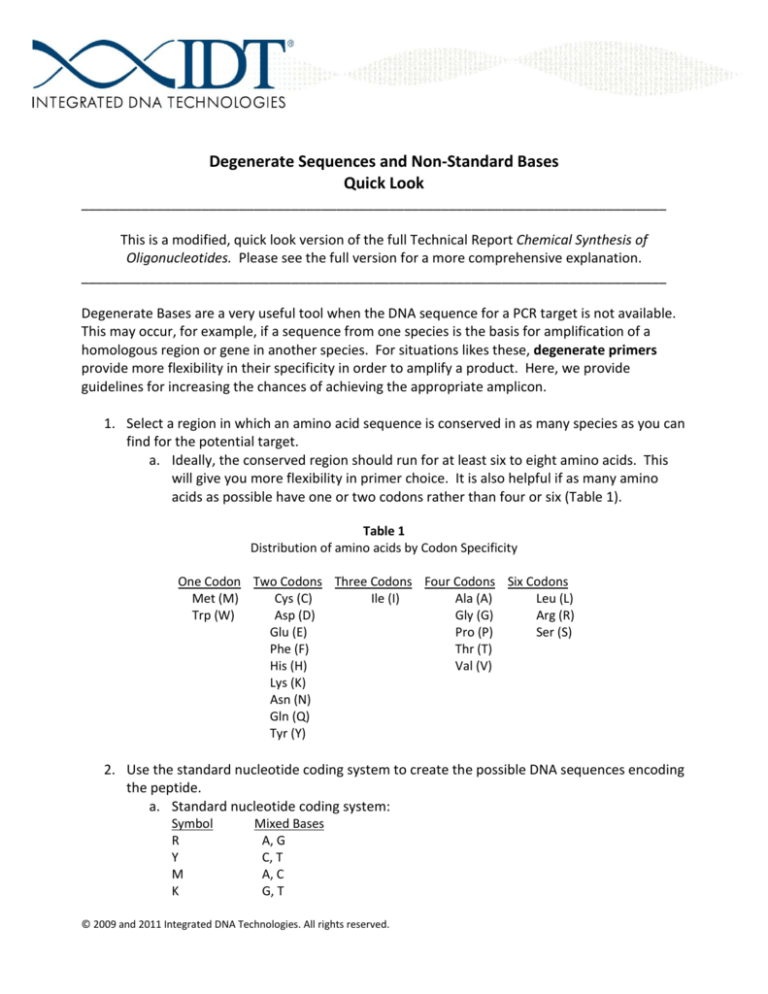
PDF] Degenerate Primer Design Using Computational Tools Computational Molecular Biology | Semantic Scholar

PANDAA intentionally violates conventional qPCR design to enable durable, mismatch-agnostic detection of highly polymorphic pathogens | Communications Biology

Table 1 from Construction of "small-intelligent" focused mutagenesis libraries using well-designed combinatorial degenerate primers. | Semantic Scholar
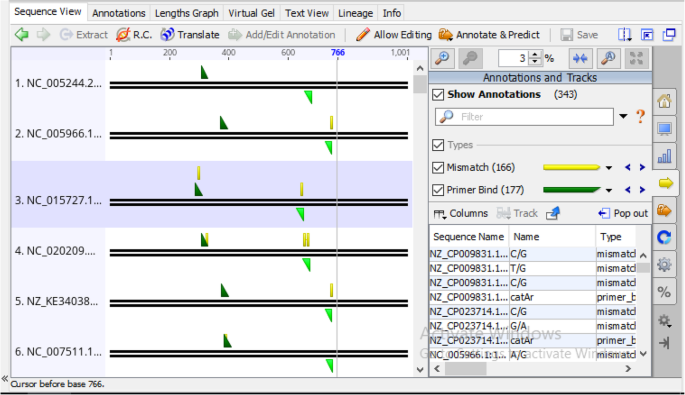
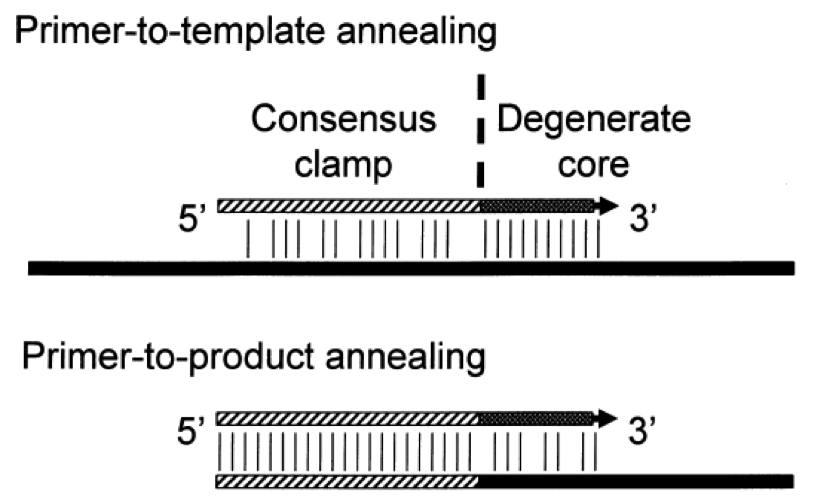
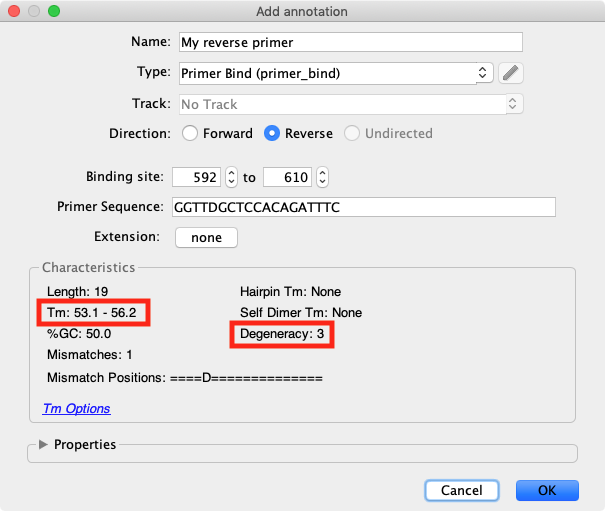
In silico design and validation of a highly degenerate primer pair: a systematic approach | Journal of Genetic Engineering and Biotechnology | Full Text